PARSNIP
PARSNIP (Protein Assessment and Ranking System for Novel Input) is a public-facing companion tool to DAIKON that supports the early stages of target evaluation. It provides a structured, evidence-based framework for scoring and comparing potential drug targets across multiple biological dimensions, and visualizes these comparisons in an interactive interface accessible to the global research community.
PARSNIP is available at: https://parsnip-56158.web.app
Overview
PARSNIP evaluates each target across three core dimensions:
- Chemical Inactivation - evidence that the target can be inhibited by a small molecule
- Genetic Inactivation - evidence from genetic studies (e.g., knockouts, CRISPRi) supporting target essentiality
- Feasibility - the practical druggability and tractability of the target
Scores across these dimensions are aggregated into a Total Inactivation score, allowing researchers to compare targets side by side and identify the most promising candidates for progression into the DAIKON pipeline.
PARSNIP Dashboard
The PARSNIP dashboard presents all evaluated targets in two complementary views:
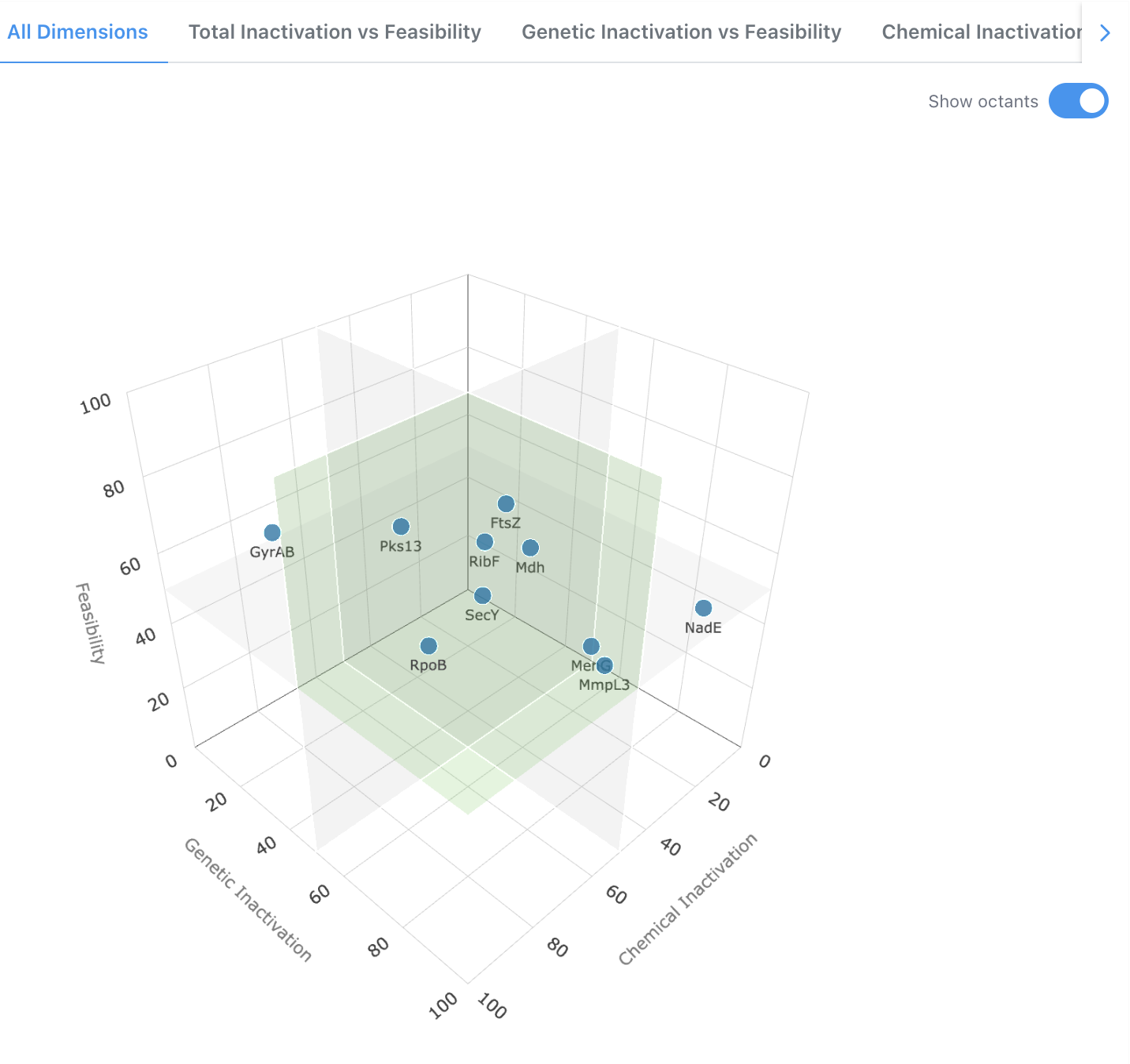
3D Scatter Plot - All Dimensions
The default view plots targets in a three-dimensional space defined by Chemical Inactivation, Genetic Inactivation, and Feasibility. An optional octant overlay can be toggled on to visually segment the space and help identify clusters of high-priority targets. Users can rotate and interact with the plot to explore target positioning across all three axes simultaneously.
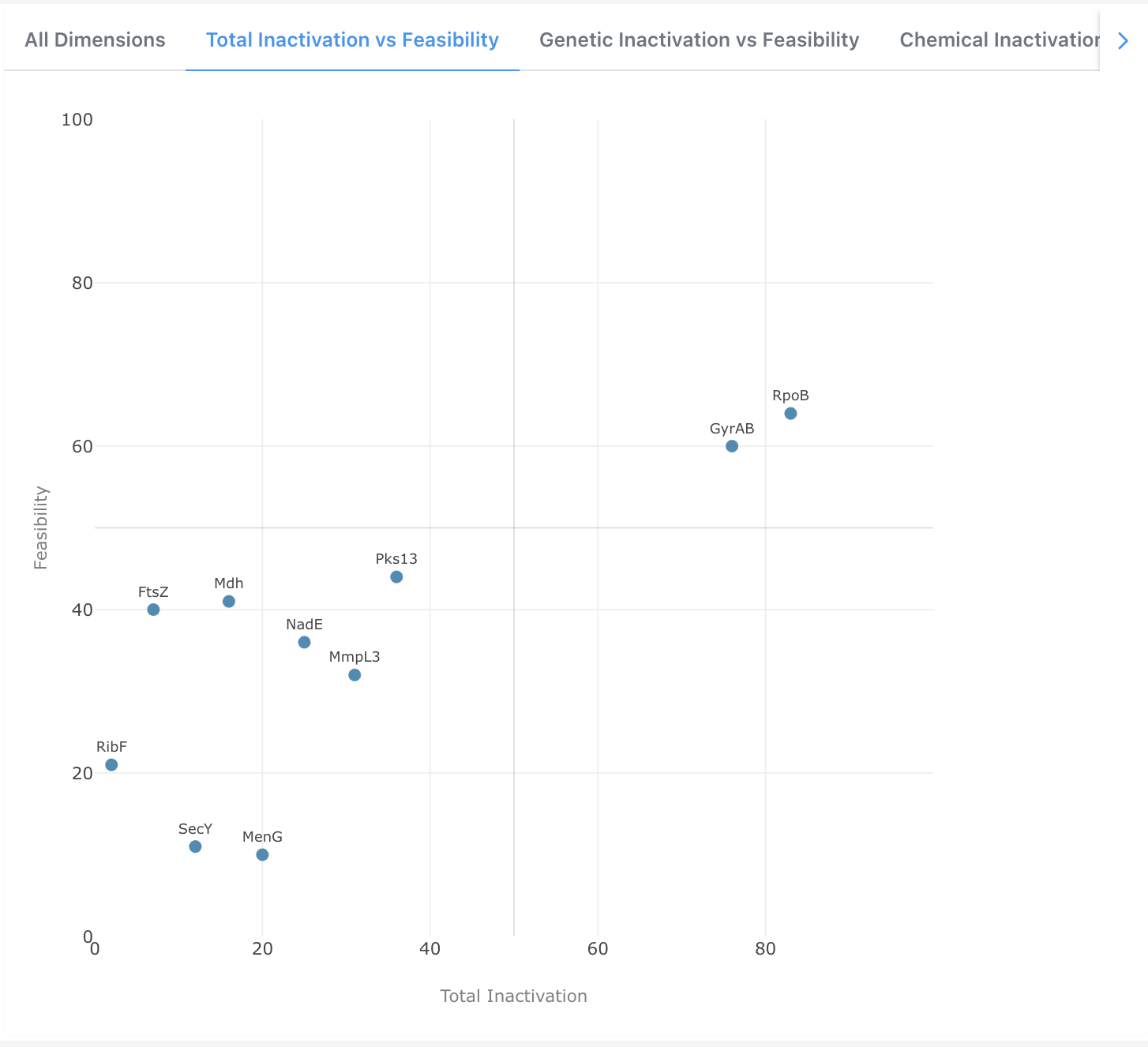
Total Inactivation vs Feasibility
A 2D scatter plot comparing each target's aggregated Total Inactivation score against its Feasibility score. Targets appearing in the upper-right quadrant combine a high level of inactivation evidence with strong druggability, making them the most attractive candidates for progression.
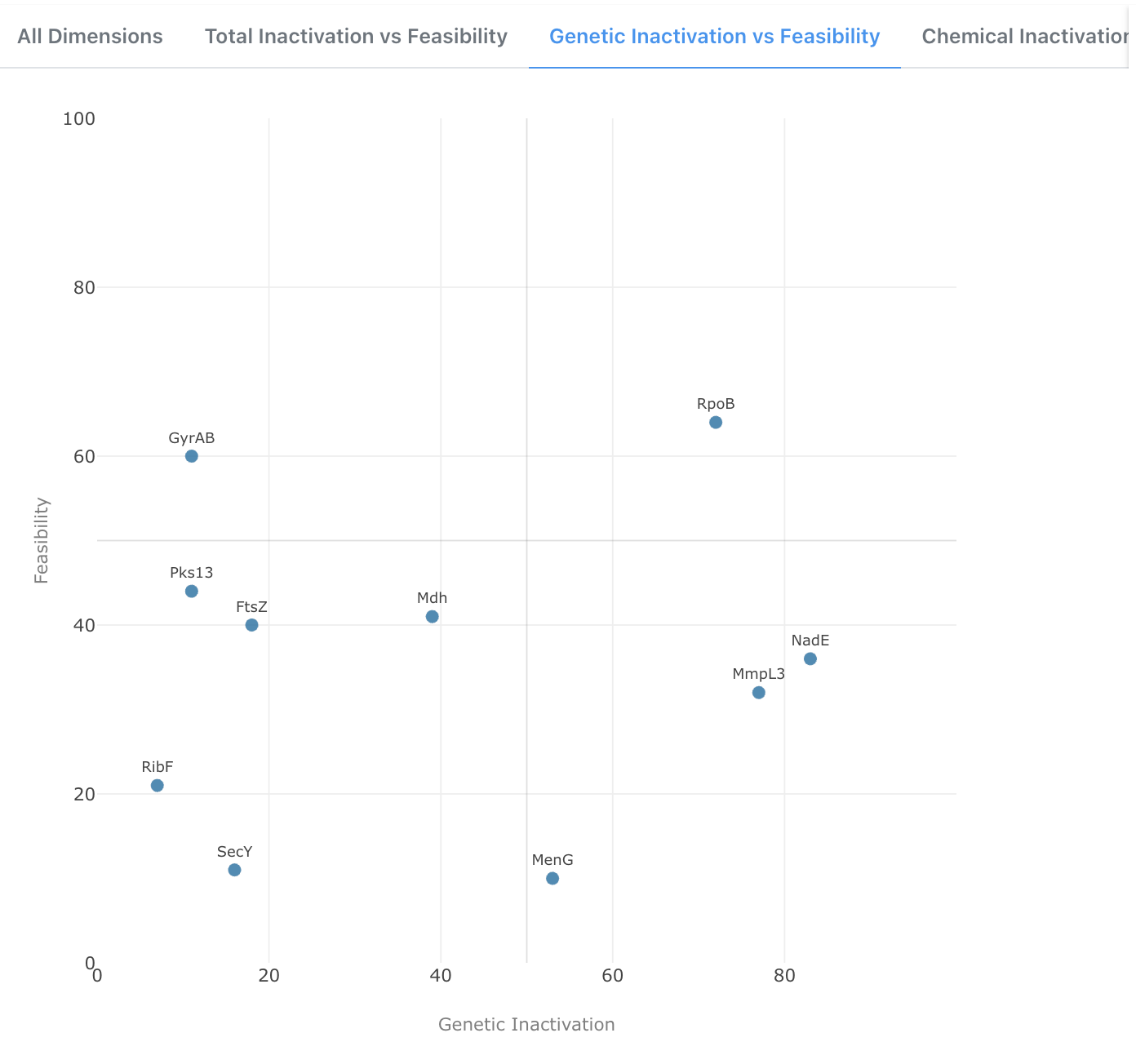
Genetic Inactivation vs Feasibility
A 2D scatter plot comparing Genetic Inactivation against Feasibility. This view is particularly useful for identifying targets with strong genetic essentiality evidence that also present a tractable path toward drug development.
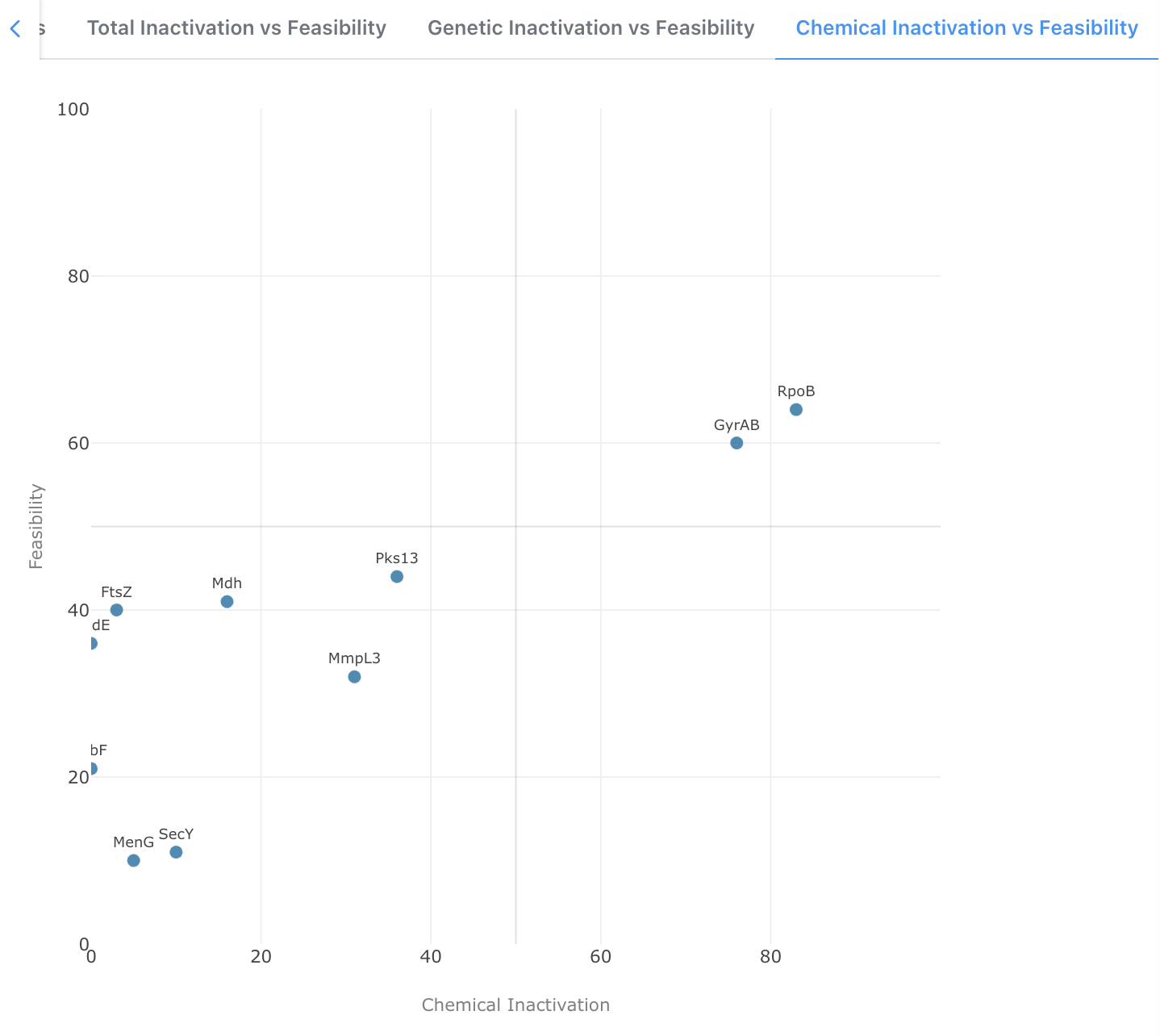
Chemical Inactivation vs Feasibility
A 2D scatter plot comparing Chemical Inactivation against Feasibility. This view highlights targets for which small-molecule inhibition has already been demonstrated, helping teams prioritize targets where chemical starting points may already exist.
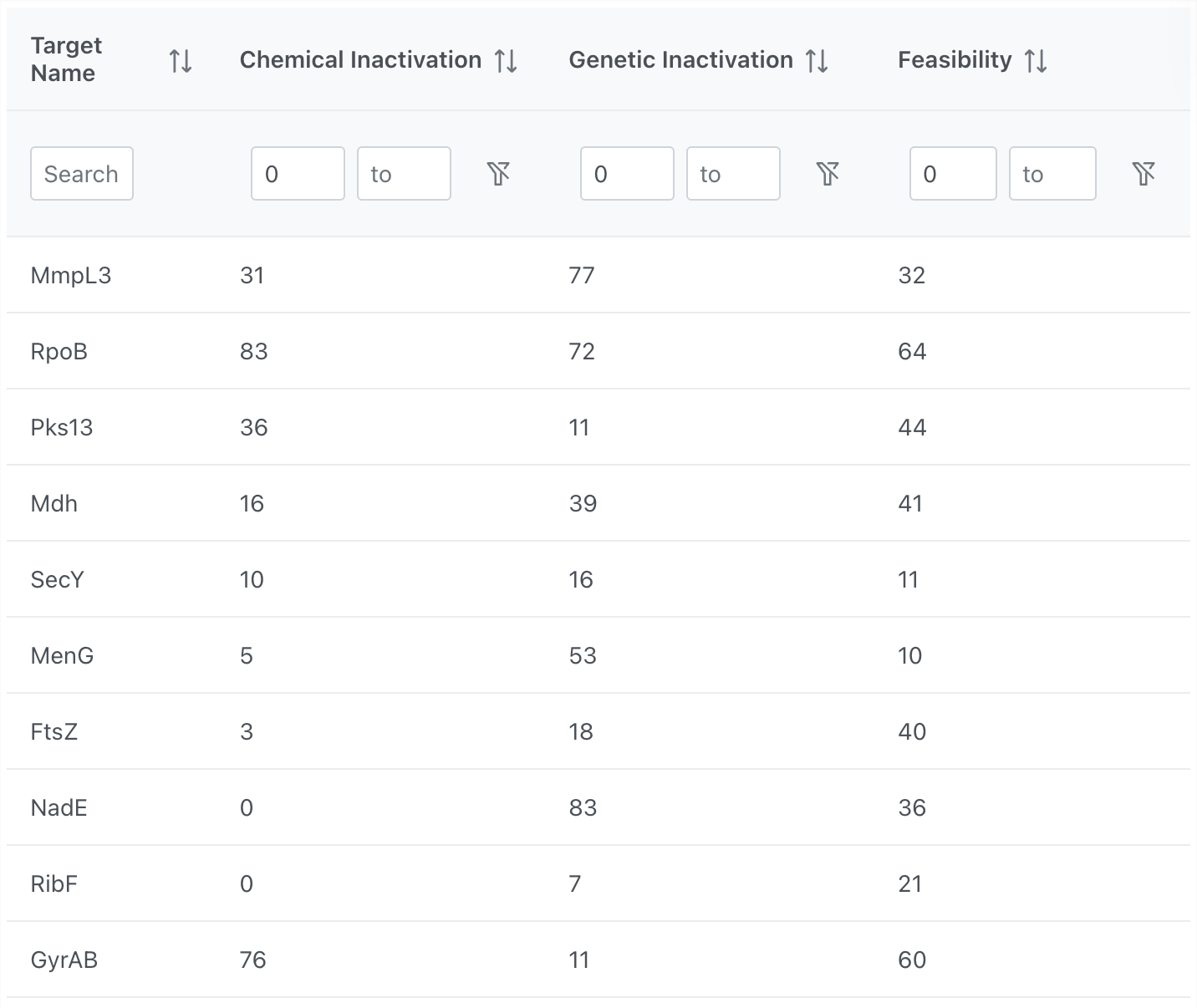
Score Table
Alongside the plot, a sortable and filterable table lists each target with its individual dimension scores. Users can:
- Search by target name
- Filter by score range for each dimension using the range inputs
- Sort any column in ascending or descending order
Target Evaluation
The Target Evaluation page allows users to score a new or existing target by completing a structured questionnaire. Scores are computed automatically across the four dimensions as answers are submitted.
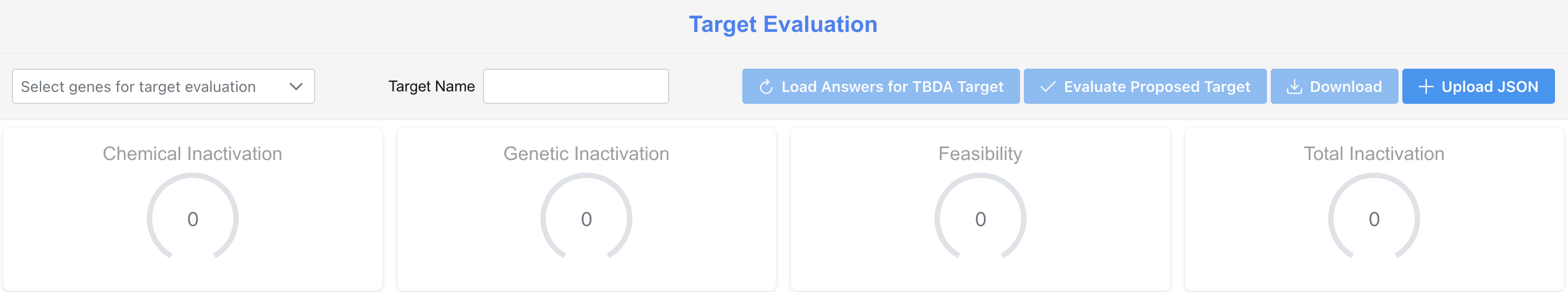
Loading an Existing Target
To load pre-populated answers for a target already evaluated, select the gene from the dropdown and click Load Answers. This will populate the questionnaire with the consensus responses.
Evaluating a Proposed Target
To evaluate a new target:
- Select the relevant gene from the gene dropdown
- Enter a Target Name
- Complete the Target Questionnaire (see below)
- Click Evaluate Proposed Target to generate scores
The four dimension gauges at the top of the page — Chemical Inactivation, Genetic Inactivation, Feasibility, and Total Inactivation — will update to reflect the computed scores.
Downloading and Uploading Results
- Download - exports the current questionnaire responses and scores as a JSON file for record-keeping or sharing
- Upload JSON - allows users to import a previously saved evaluation to resume or review responses.
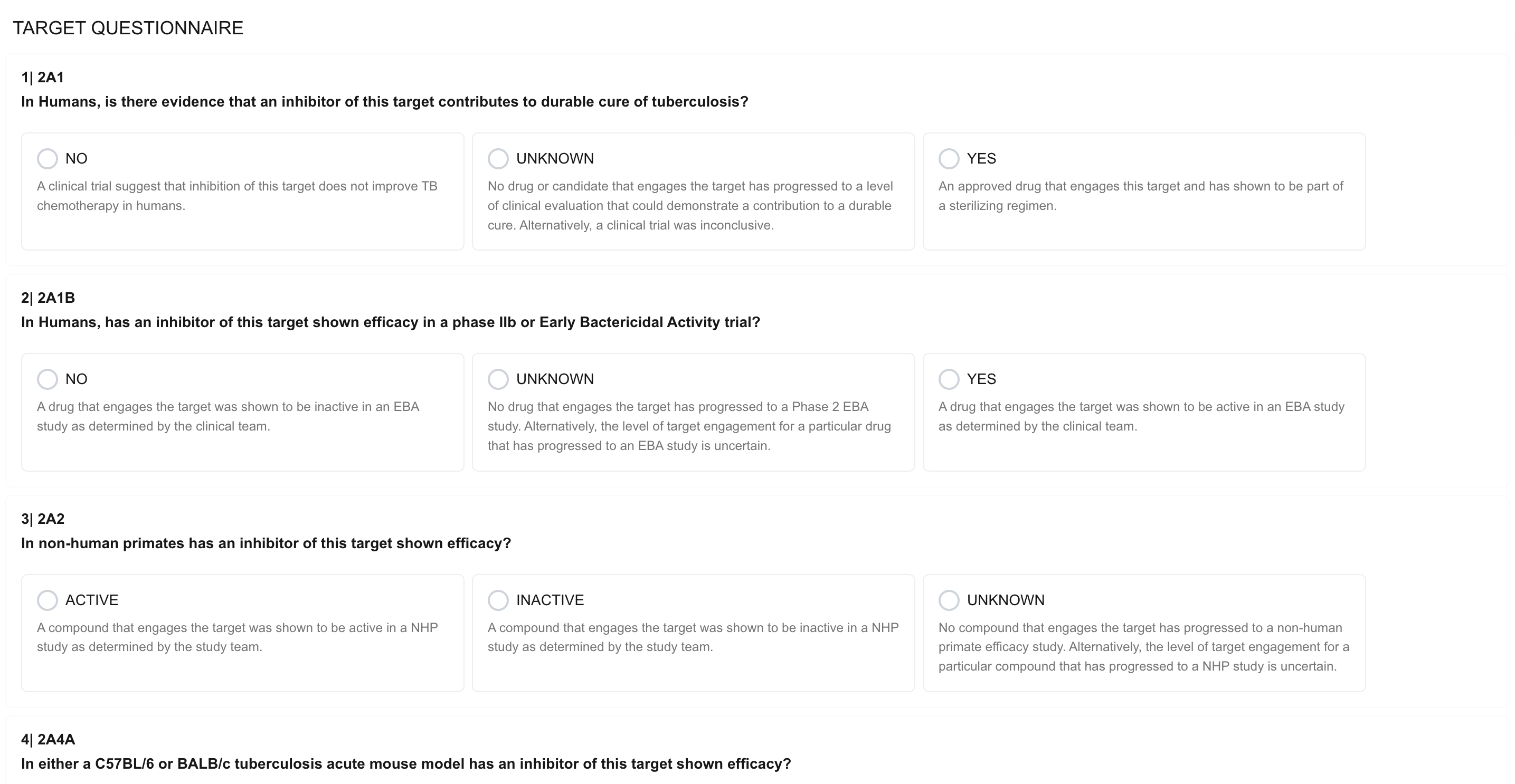
Target Questionnaire
The Target Questionnaire is the core scoring instrument in PARSNIP. It presents a series of structured questions across the Chemical Inactivation, Genetic Inactivation, and Feasibility dimensions. Each question offers clearly defined response options (e.g., YES / NO / UNKNOWN, or ACTIVE / INACTIVE / UNKNOWN), with descriptive guidance provided for each choice to ensure consistent interpretation across users.
Example questions include:
- In humans, is there evidence that an inhibitor of this target contributes to durable cure of tuberculosis?
- In humans, has an inhibitor of this target shown efficacy in a Phase IIb or Early Bactericidal Activity trial?
- In non-human primates, has an inhibitor of this target shown efficacy?
Responses to each question contribute to the overall dimension scores, which are displayed in real time as the questionnaire is completed.
Relationship to DAIKON
PARSNIP operationalizes the Target Prioritization Module described in the DAIKON framework. Within DAIKON, this module functions as an abstract layer that accepts quantitative or qualitative scoring inputs to promote high-priority proteins from the gene pool into the active target pipeline. PARSNIP makes this layer publicly accessible, enabling researchers outside the TBDA consortium to apply the same prioritization methodology and contribute evaluations of proposed targets.